1. A complete list of web services.
TOOLS PAGE:The main function of this page is to predict the interaction between drugs and targets, and to further predict the affinity and interaction sites based on the predicted interaction values. The DTI model we use is MGraphDTI,which is from the paper "MGraphDTA: deep multiscale graph neural network for explainable drug–target binding affinity prediction", published in the journal Chemical Science in March 2022. In this study, we used the benchmark datasets Human and C. elegans for a binary classification task. Finally, the model achieved performance on two different datasets with Precision: 0.955(0.005), Recall: 0.956(0.003), AUC: 0.983(0.003) and Precision: 0.980(0.004), Recall: 0.967(0.005), AUC: 0.991(0.001). You can view the paper through the following link: https://doi.org/10.1039/D1SC05180F.For the DTA model, we adopted our research group's MMDTA model, which is from the paper "MMDTA: A Multimodal Deep Model for Drug-Target Affinity with a Hybrid Fusion Strategy", published in JCIM in August 23, 2023. The model's dataset uses the core set data of pdbbindv2016. You can view the paper through the following link: https://doi.org/10.1021/acs.jcim.3c00866.The DTS model uses an integrated model combining graph convolutional network (GCN) and graph attention network (GAT) for predicting the binding sites of drug-target interactions, as described in the paper "Hybrid protein-ligand binding residue prediction with protein language models: Does the structure matter?". You can view the paper through the following link: https://doi.org/10.1101/2023.08.11.553028. You can enter our tool interface through this link: http://39.106.7.26:8000/Drug-Online/. Detailed usage of the tool will be explained later in the document!!!
API PAGE:The API interface primarily introduces what an API interface is and its purpose. It also allows us to include our pre-written script code within the page, enabling direct usage of the script code. By replacing the file addresses with our own, we can directly access our server tool and obtain the output results.You can view it through the following link: http://39.106.7.26:8000/api/ .
2. Demonstration of web server functionalities example.
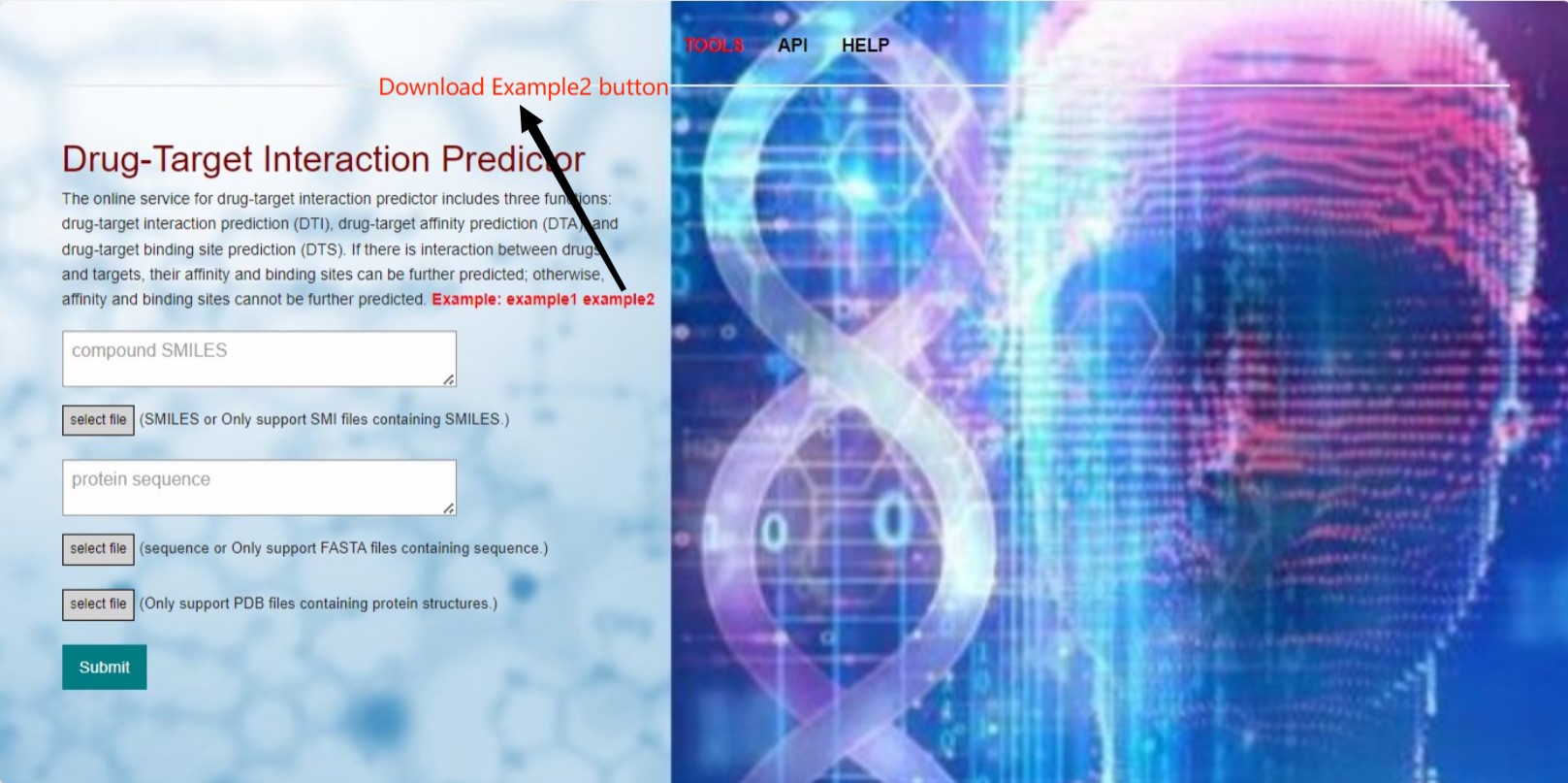
The main function of the web server is to perform calculations for drug-target interactions. As mentioned before, we will use three different models to calculate different metrics. However, when there is no interaction between the drug and target, i.e., when the DTI prediction for binary classification is 0, we will not proceed with the prediction of affinity and binding sites. Only when there is a direct interaction between the drug and target, i.e., when the prediction value is 1, we will perform predictions for DTA affinity and DTS binding sites.The example provided includes data for these two different scenarios, which correspond to two different results. We will now describe in detail the steps involved in running these two scenarios:(The following image shows some operation buttons and information in our tool interface. The tool allows input of compound sequence and protein sequence files as well as corresponding compound/protein sequences. However, protein structures can only be input as pdb files.)

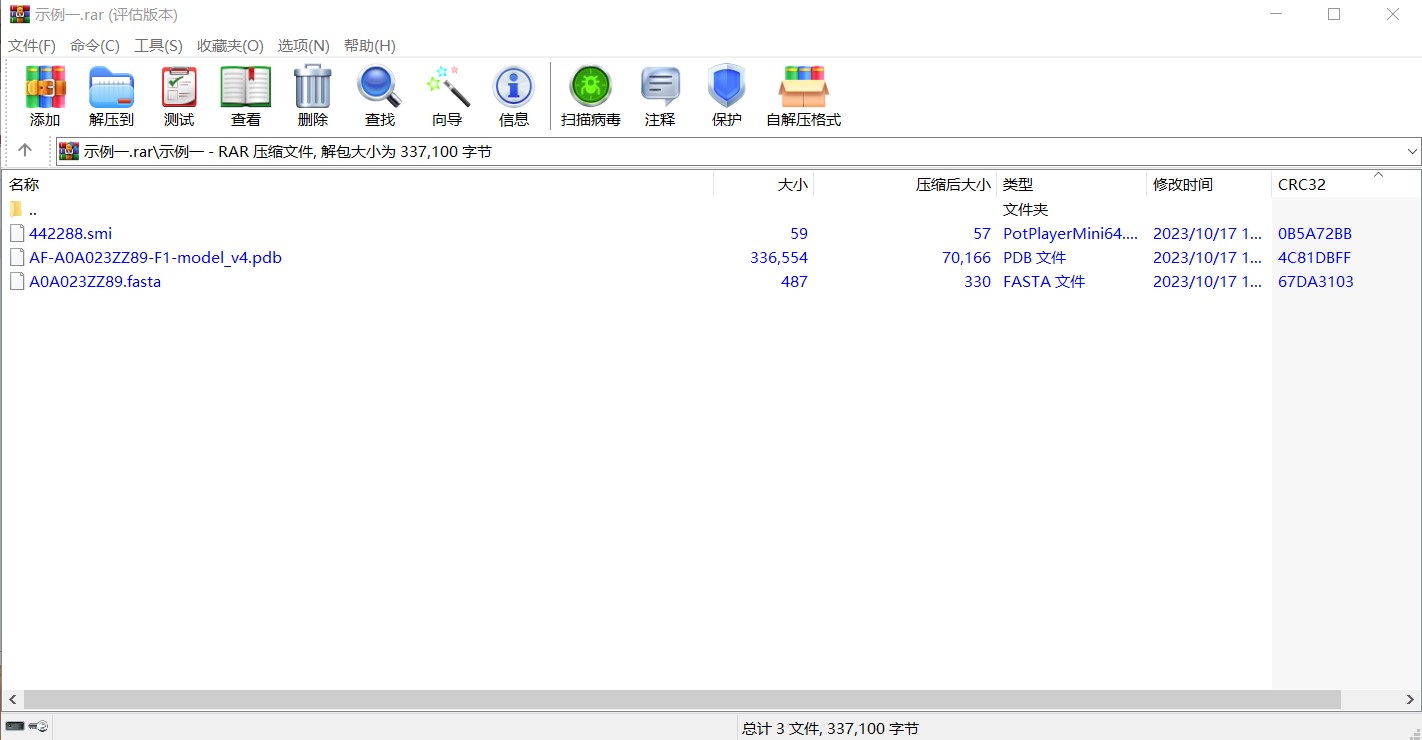
Example 1: First, download the compressed file for example 1 by clicking on the download button. The compressed file contains three files, including compound sequence file and protein sequence and structure files. As shown in the image below:
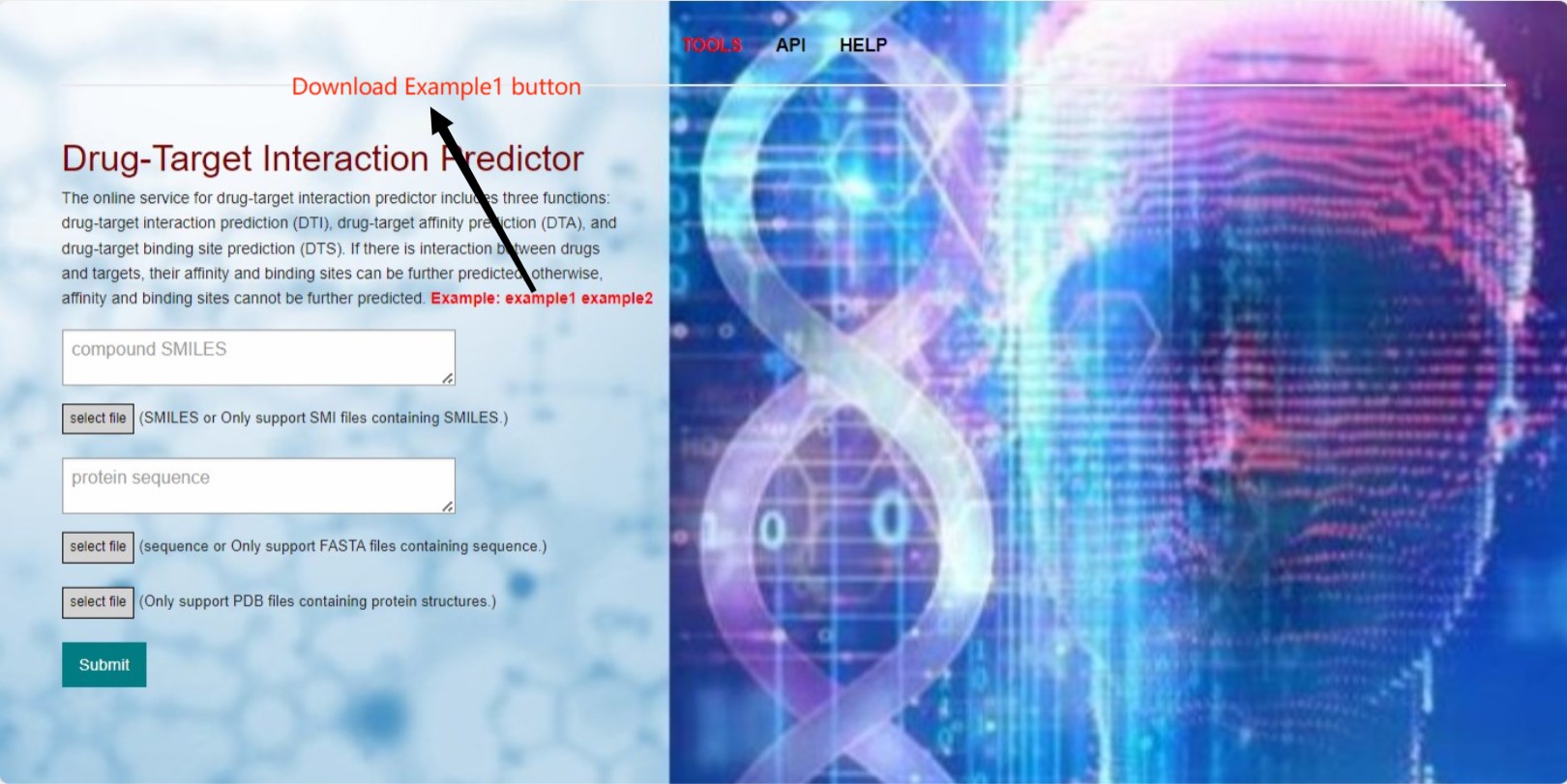
Next, load the corresponding compound sequence file, protein sequence file, and protein structure file into the submission server tool.As shown in the image below:

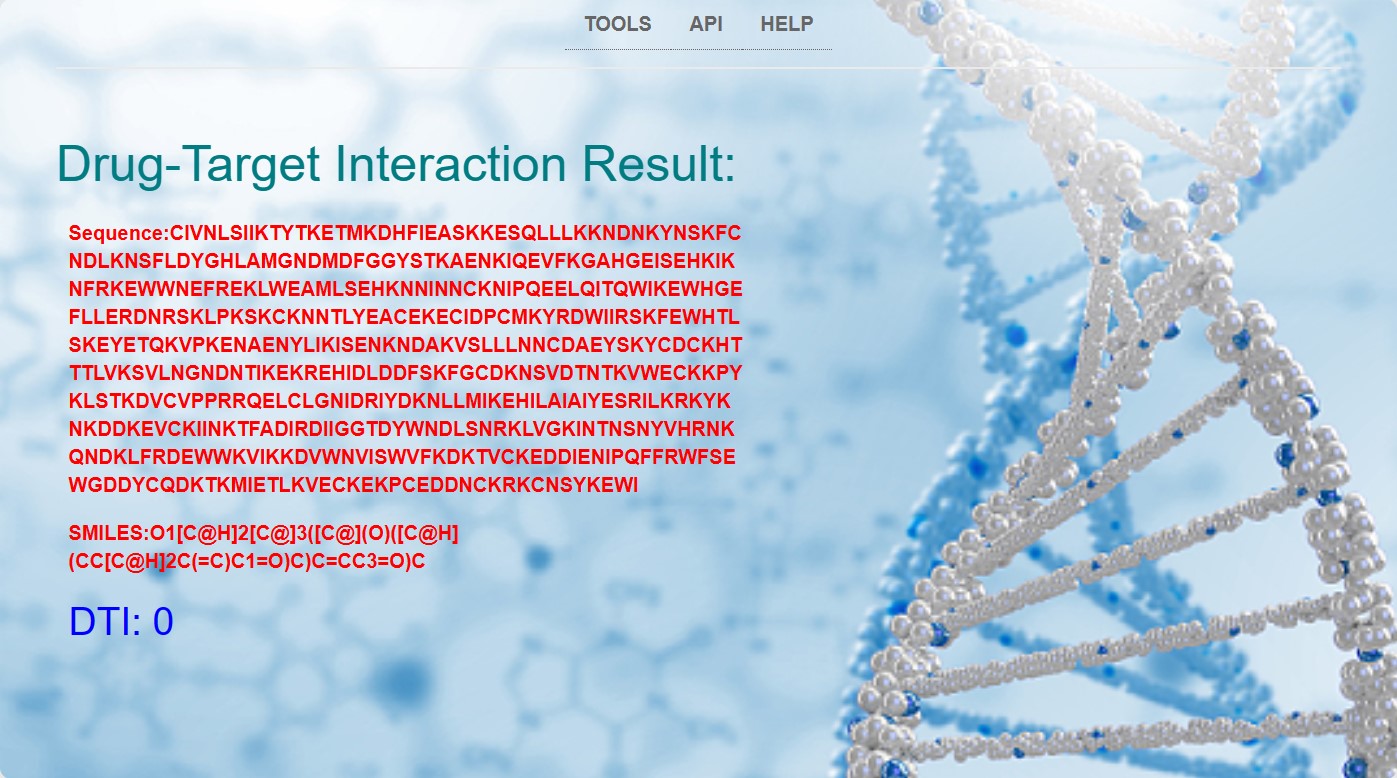
If the input drug-target interaction (DTI) prediction for both the compound and protein in Example 1 is 0, then the result page will directly output the sequences of the protein and compound, as well as the DTI prediction value!The result of Example 1 is shown in the image below:(The results of Example 1 can also be viewed through the following link: http://39.106.7.26:8000/example1/ !!!)
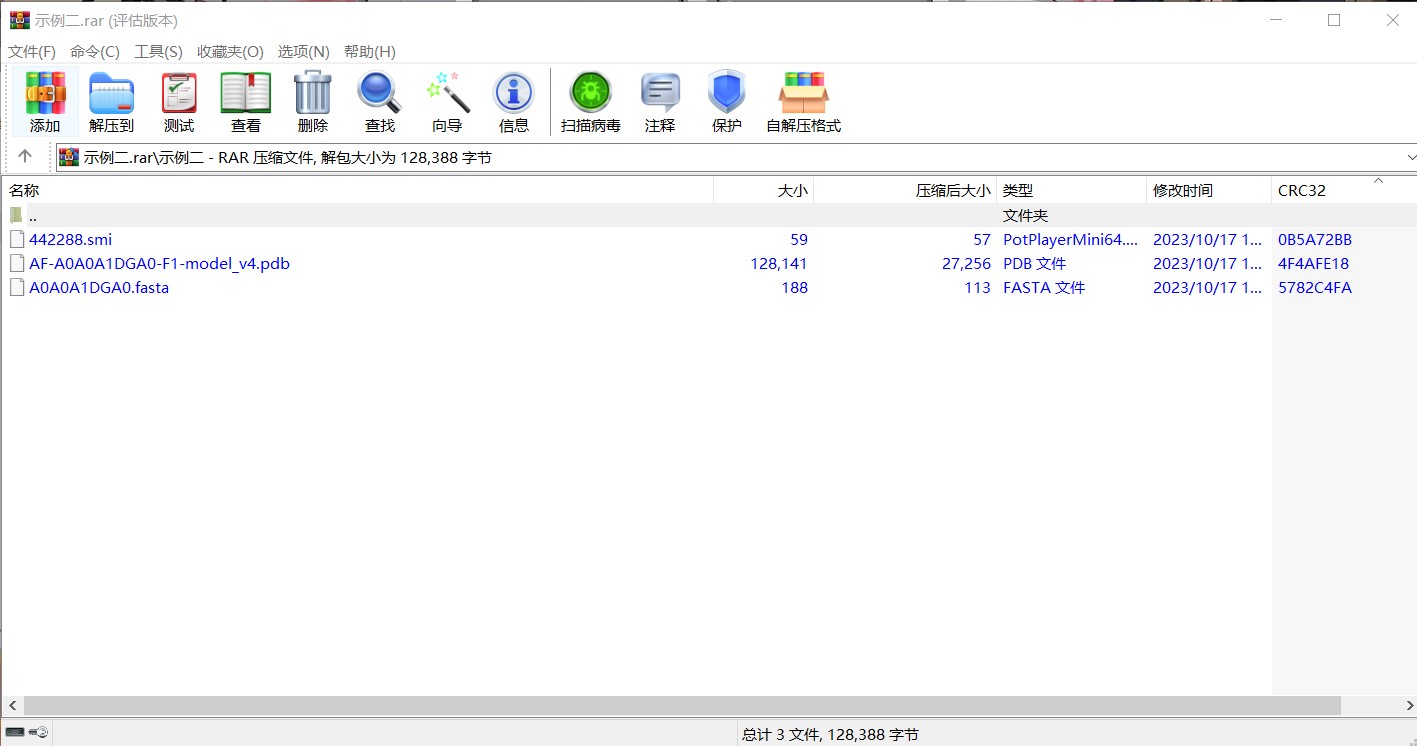
Example 2:First, download the compressed file for example 2 by clicking on the download button. The compressed file contains three files, including compound sequence file and protein sequence and structure files.As shown in the image below:
Next, load the corresponding compound sequence file, protein sequence file, and protein structure file into the submission server tool.As shown in the image below:

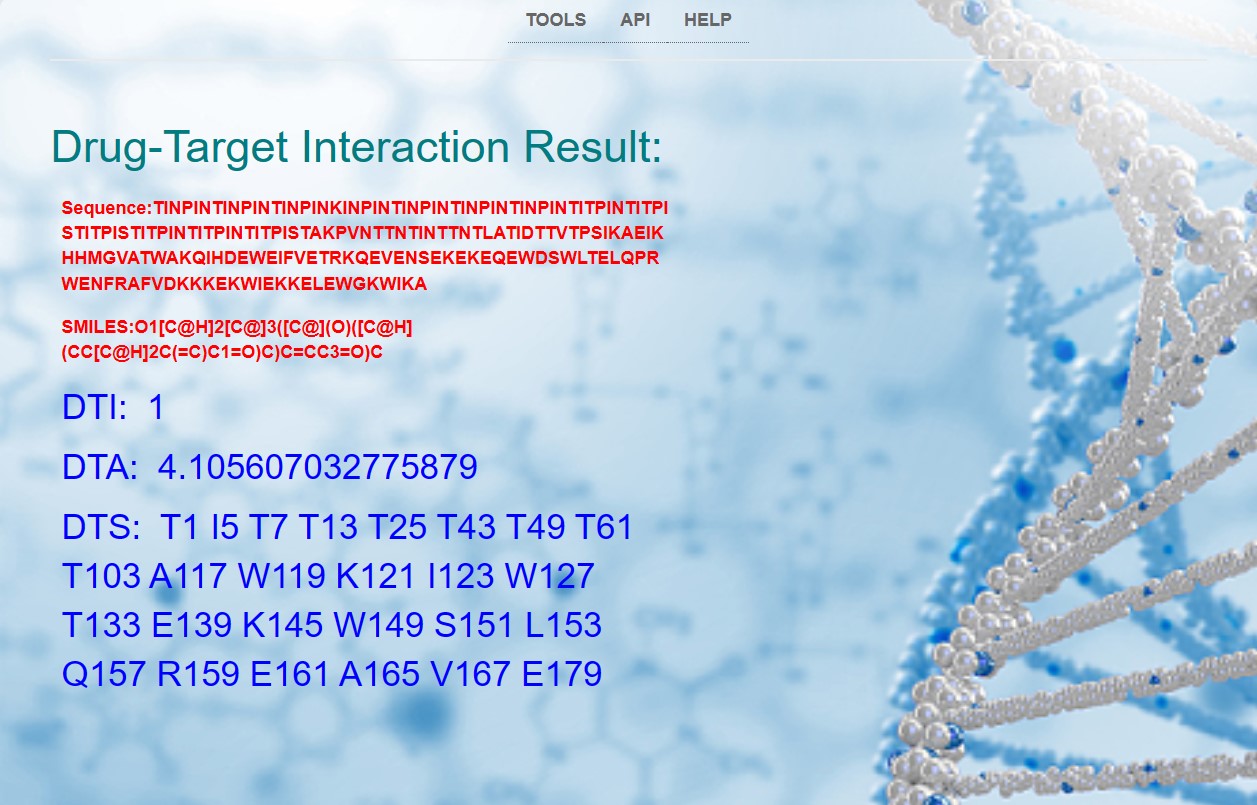
If the input drug-target interaction (DTI) prediction for both the compound and protein in Example 2 is 1, then the result page will directly output the sequences of the protein and compound, as well as the predictions for DTI, DTA, and DTS!The result of Example 2 is shown in the image below:(The results of Example 2 can also be viewed through the following link: http://39.106.7.26:8000/example2/ !!!)